Imagine you are a bacterium, roughly 1/1,700,000 of your current size, residing in your own human body’s gut. You live in a diverse community, the “microbiome,” teeming with other bacteria: friendly neighbors who live next door, some ne’erdowells who occasionally vandalize the town, and your neighborhood cops who try to keep everything in check. The overall health of this community directly affects the health of the person that you live in.
As a bacterium, you are affected by what your host eats, what chemicals are in their environment, even what your host’s mother ate when she was pregnant. As is typical of bacteria, you often swap genes with your neighbors. On occasion, your host gets sick. She takes an antibiotic and many individuals in the community are killed, including both vandals and cops, but you are lucky to have picked up some genes that allow you to survive. Over time, the vandals also pick up some antibiotic-resistant genes. If there aren’t enough cops or other bacteria around to compete with these hardened bad guys and keep their numbers down, they can dig in and cause a potentially fatal antibiotic-resistant disease.
In May 2014, the World Health Organization reported that antibiotic resistance is on the rise, with no signs of slowing down. Strains of E coli responsible for causing urinary tract and blood infections, is becoming unfazed by front-line therapies. Resistant forms of pneumonia, gonorrhea, and tuberculosis are becoming prevalent, as well.
The rate at which resistance to antibiotics is developing for a range of infections far exceeds that at which new antibiotics are being developed. In fact, the development of antibiotics has plummeted significantly: between 1980 and 2000, 51 new antibiotics were FDA-approved; only 10 have been approved since 2000. And some of the new ones are not exactly widely accessible: Dalvance, approved in May 2014, comes at a hefty price tag of nearly $3,000 a dose.
“We can think about the [health of a person’s] microbiome like global warming. The microbiome is the same way. When you first start dumping antibiotics, that’s like releasing carbon dioxide into the atmosphere. It may not do much the first time, but every time you take an antibiotic, it adds up.”
Henry Chambers, the head of the Antibacterial Resistance Leadership Group at University of California, San Francisco, says that developing more antibiotics is not the answer. “Not to be dismissive, but making more antibiotics will only cause more problems, since bacteria will figure out how to be more resistant,” he says.
Chambers and an increasing number of infectious-disease experts say the real problem is over-prescription of antibiotics, especially to young patients. “We can think about the [health of a person’s] microbiome like global warming,” says Chambers. “If someone told you that by starting your car, you would be releasing fossil fuels into the environment, you’d contribute to that problem every time you use your car. It’s hardly detectable at first, but when spread out over a large population, you see its effects. The microbiome is the same way. When you first start dumping antibiotics, that’s like releasing carbon dioxide into the atmosphere. It may not do much the first time, but every time you take an antibiotic, it adds up.”
Researchers say one solution is to use less antibiotics but use them in a smarter way. To do that, we need to be able to determine exactly what kind of bacteria is causing each infection. Chambers says that better diagnostics could “revolutionize the field.”
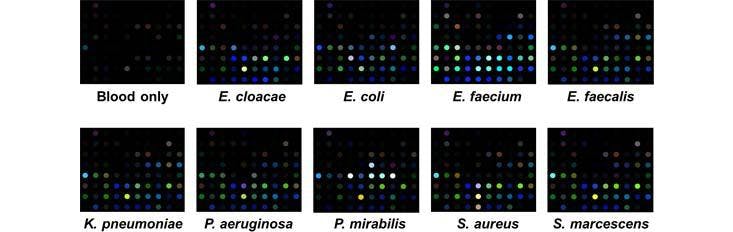
Specific Technologies, a biotechnology startup, has been developing an inexpensive and reliable sensor called SpecID as a diagnostic tool. This “artificial nose” includes an array of spots, each spot being a single dye that changes color when exposed some chemical that bacteria produce. For example, thiols, one of the many types of compounds emitted by bacteria, can be detected by a handful of dyes. The array is fixed under the cap of a vial filled with a patient’s blood. Any bacteria living in the blood secrete certain chemicals, and each species produces a mixture of metabolites specific to their species. The combination of chemicals released by each strain of bacteria generates a distinctive colored pattern.
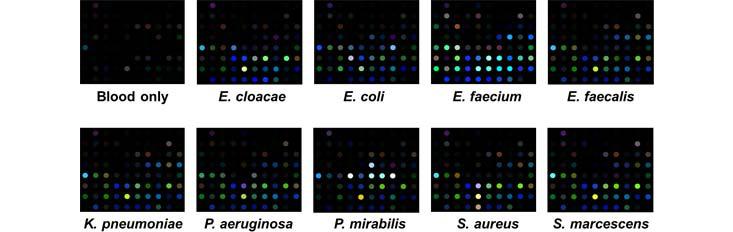
In lab experiments, Specific Technologies found that clinicians could quickly determine whether an infection was bacterial or viral. (If the array doesn’t light up at all, the infection is thought to be viral.) Next, they can match the developing array pattern with known patterns in a computer reference system to distinguish 18 important infectious bacteria and 60 strains with over 90% accuracy. The company says it can even determine whether certain bacteria are resistant or sensitive to antibiotics; knowing this could save the doctor and patient from the risk of taking a drug that will not be effective.
Currently the process of determining the cause of infection is laborious: Samples must be taken from the clinic into a lab, where cultures are slowly grown. It takes 12 hours, on average, to establish whether an infection is viral bacterial, and an additional 24 hours or so to determine a bacterial strain. SpecID cuts those times down to around two hours and 15 hours, respectively, according to Specific. That acceleration could be life-saving for patients who present with sepsis, a life-threatening condition that can arise from blood infection. Paul Rhodes, CEO of Specific Technology, hopes that SpecID will enter clinical trials in early 2015.
While antibiotics act as a bomb, indiscriminately killing both bad and good bacteria and potentially leading to an imbalanced microbiome, doctors are also working on ways to out-compete pathogens, by fostering healthy microbiomes in patients. Listed as an “urgent threat” by the CDC, Clostridium difficile infections are resistant to antibiotics but are often effectively relieved by fecal transplants, where the patient’s compromised microbiome is replenished with a healthy one. Donor stool is diluted with saline before being injected to the patient’s gastrointestinal tract. The effectiveness of this procedure for the treatment of a devastating, antibiotic-resistant condition illuminates the importance of maintaining a balanced microbiome.
Certain herbs, like those used in traditional Chinese medicine, may be another, less destructive way to keep harmful bacteria away. Some herbs, like berberine, and andrographis, have well-documented antimicrobial effects. There are also a number of other herbs which indirectly influence the microbiome by activating parts of the patient’s immune system. The immune system, in turn, tunes the host microbiome, making it better at keeping harmful bacteria out. Crucially, it seems that herbal medicine is less likely than antibiotics to generate resistance: Many herbs act on the immune system rather than the bacteria themselves, and the herbs that do kill bacteria are often used in combination (the combined activity is less likely to leave survivors that can withstand one chemical).
All of this isn’t to say that we should abandon antibiotics. Even Chinese-medicine doctors Elizabeth Stephens calls for them when patients come in with “oozing sores on their throat” from an upper respiratory infection, for instance. But “unless there’s an urgent need for antibiotics, we should try and resort to gentler means,” she says.
As our bacteria find more clever ways to evade death-by-antibiotics, humans should reassess how we engage in this challenge. If we can better understand the body as an ecosystem and health on a holistic level, we will take one step closer to outsmarting the bacteria that plague us.
Wudan Yan (@wudanyan) is a freelance science journalist based in New York City. She is also the science correspondent / science editor for HippoReads.